In attendance:
Mark, Eddie, Wendy, Richard, Ivan, Andreas, Paul, Martin, Mylah
We started with jokes about the danger of having the core Moby developers in the same room of a brick hospital built on top of a geological fault…
Issues that came up in introductory presentations:
Atrtribution of data to providers
– set logo? new methods…
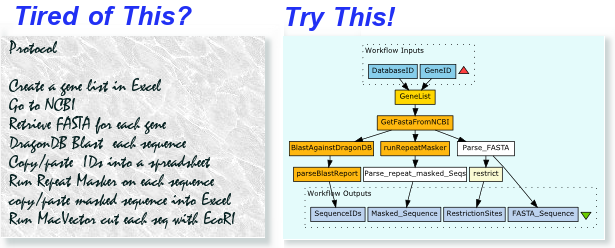
Should the Object ontology have multiple representations for any given concept?
– e.g. FASTA and DNA Sequence are redundant…
– we need to find a way to encourage object re-use
– we need a “rule-base” to use the SeaHawk transformation rules rather than registering a new Moby object; or do we still need to register a new object?
– how much of this complexity should we expose the developers to?
In what order do we introduce the concepts of Moby to newcomers? How do we explain “the point” to newbies? The idea of data-type re-use, for example.
There’s no “one ring/protocol to rule them all”
– TAPIR/DIGR/BioCASE
– SSWAP
– Moby
– Gramene format
Ivan apologizes for his English 🙂
– it really isn’t as bad as he claims it is!
Need to support large datasets
Need to support document-encoded rather than RPC
– Java libraries no longer support RPC SOAP
– Perl library doesn’t support anything OTHER than RPC SOAP
– this is clearly a problem!
Support “template datatypes” to contain other kinds of objects