Table of Contents
Introduction
We updated the BioMoby plugin so that we could add functionality that we believed was missing.
In the past, users of the plugin were forced to start every Moby based workflow with the root Moby
datatype. In other words, you were able to utilize a local widget called 'create_moby_data', however,
the Moby data really was just the root datatype in disguise.In addition, in Taverna releases prior to version 1.2, the BioMoby services were always limited to one
input and one output. This worked for many of the Moby services, but for those wishing to create a
workflow that initally utilized a service with more inputs, that option just wasn't available.In between Taverna releases, you can always get the most up-to-date version of BioMoby plugin by
visiting the biomoby.org or building your own from the Taverna CVS.For example workflows that utilize BioMoby services, please click here. If you have a workflow
that you would like to share with others, please submit it to me and I will make it publicaly available.
New Features
The updated BioMoby plugin contains many new features. Some of these features include the following:
- Secondary parameters are now supported.
- New ways to parse service outputs.
- Ability to add datatype parsers from the 'Advanced Model Explorer'
- Caching of Services/Datatypes from different registries that should aide in improved loading times.
- Service notes and service exceptions are no longer removed from messages so that you
can view them.- A new output port for Moby collections called 'AsSimples'. This port breaks apart
Moby collections into a Taverna list of 'Simples' so that a downstream service can
properly utilize the items in the collection.- Persistance of the mobyData elements attribute queryID. This will allow better
tracking of inputs and outputs from Moby services.- The ability to search for Moby services using the outputs from another Moby service.
When searching for services, namespaces are respected.- Removed support for Moby services that are not registered correctly, ie ones that
contain names with "_ANON_" and "(Collection -'MobyCollection')". These services are
incorrectly registered with Mobycentral and, in most cases, aren't working correctly.- When using the context menu in the 'Moby Service Details' window, input datatypes
added to the workflow are automatically linked to their respective service.
Tutorial
Creating a BioMoby Scavenger
Creating a new BioMoby scavenger has never been easier! In fact, if you are familar with Taverna,
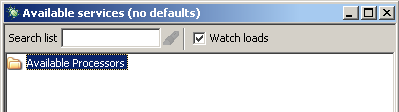
then adding a BioMoby scavenger is just like adding any other scavenger.Starting from the 'Available Services' window, you will need to context click the folder labeled
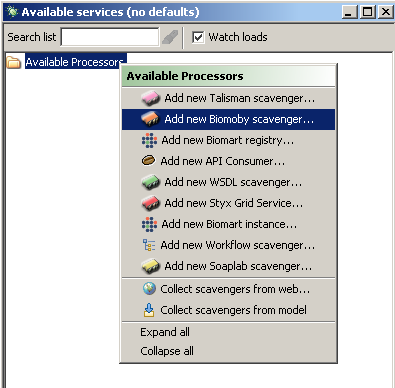
'Available Processors'. Below is a image illustrating the 'Available Services' window.
Performing a context click on the selected folder reveals the following menu. Notice how
we chose to add new BioMoby scavenger.
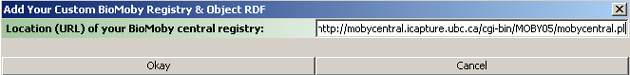
Upon making our selection, we will be prompted to enter certain information that will allow Taverna
to create our scavenger. The information that we need to provide is the URL of the BioMoby
central registry that we would like to use.
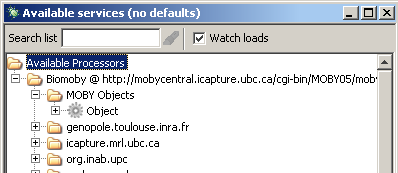
Once the information has been inputted into the text box, the Moby services and datatypes will now be available
as processors in the 'Available Services' window.
If you would like to work with your own custom Mobycentral registry and not that of the default, http://mobycentral.icapture.ubc.ca/cgi-bin/MOBY05/mobycentral.pl, then it is recommended that you edit the /taverna-workbench-1.3/conf/mygrid.properties file. In this file, modify the value for the property taverna.defaultbiomoby replacing the URL with that of your custom registry.
Creating a workflow
Workflow used in this tutorial may be downloaded here.
Workflow input used in this tutorial may be downloaded here.Creating a workflow with the new BioMoby plugin is as easy as it was using the previous plugin. There are, of course,
a few differences. When creating workflows, it is no longer necessary to use the local widget 'create_moby_data'.
When you are ready to instantiate a Moby datatype, all that is necessary is for you to choose the datatype from the list
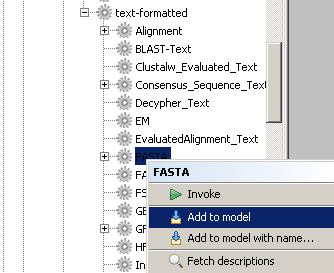
of 'Available Processors' under the 'Moby Objects' folder.As an example, we will add the Moby datatype called FASTA to our workflow. In order to do this, we must
navigate the datatype ontology to find FASTA. We can find FASTA by traversing the 'Moby Objects'
tree in the following order: Object, text-plain, text-formatted, and finally FASTA.
Now that we have found the processor that we were looking for, let's add it to the workflow.
'FASTA' has now been added to the workflow diagram. To reveal what inputs and outputs this processor
utilizes, we need to ask Taverna to show us all of the ports in the workflow diagram.
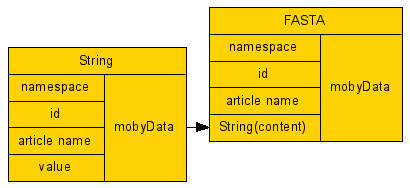
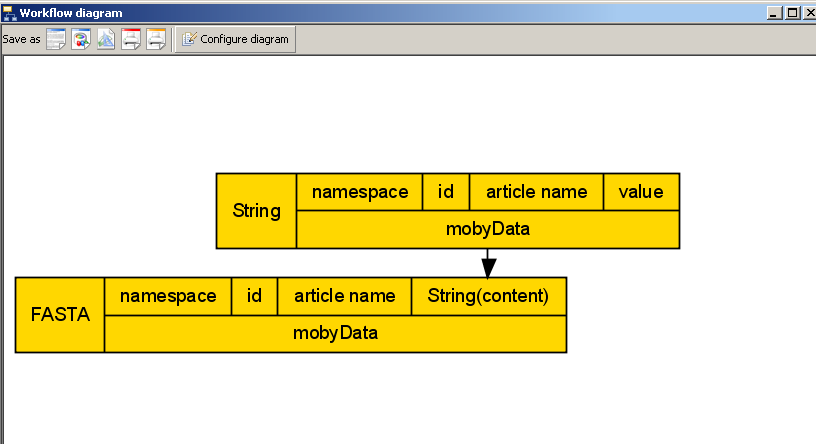
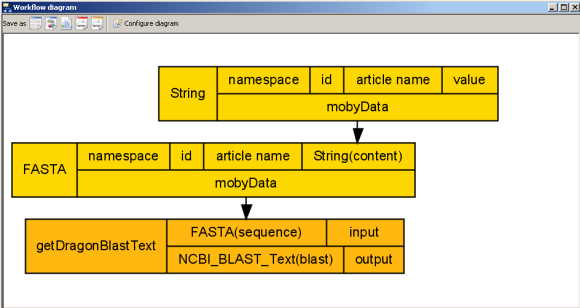
Notice how 'FASTA' takes as input a namespace, an id, an article name, and 1 other Moby datatype - String.
Since 'String' is a Moby datatype contained within a 'FASTA', it has been conveniently
added to the workflow and linked appropriately.Our workflow diagram looks something like the following:
A complex BioMoby datatype. 'FASTA', and its subcomponent, String, have now been added to our workflow.
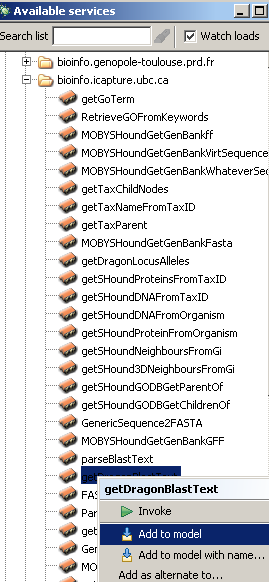
Next we will add a BioMoby service processor that handles our complex datatype.We will add the service 'getDragonBlastText' from the authority 'bioinfo.icapture.ubc.ca'. This service runs the blast tool on the input
FASTA sequence . From the list of processors, we will navigate to bioinfo.icapture.ubc.ca and then choose the service that we are
interested in, i.e. 'getDragonBlastText'.
The next thing to do is to connect the input of the 'getDragonBlastText' processor to the output of 'FASTA'. It should be noted
that there are 2 inputs to the processor 'getDragonBlastText': 'FASTA(sequence)' and 'input'. The port labelled 'input' serves to
support workflows created prior to Taverna 1.2 and should no longer be used when you create new workflows with this plugin.
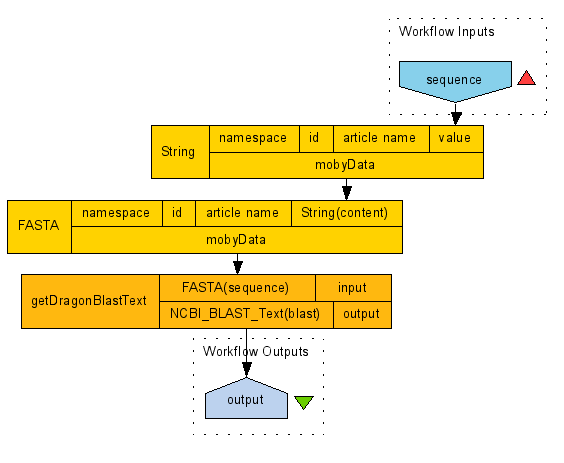
For our workflow, we will connect the complex datatype 'FASTA' to the input port labeled
'FASTA(sequence)' of the 'getDragonBlastText' processor.
We are almost done! The only things that are missing now are the workflow inputs and the workflow outputs. We will create
the following workflow inputs:
- sequence - this input will connect to the 'value' input port of 'String'
We will also create a workflow output called 'output'. This workflow output will be connected to the
output port, 'NCBI_BLAST_Text(blast)' of the processor 'getDragonBlastText'.Once these connections have been made, you should have something similar to the image below. Note that the image is only showing
those ports that are bound to inputs or outputs.
So there we have a completed workflow based on BioMoby Services and datatypes. If you wish to run this
workflow, some suitable values for the workflow inputs are as follows:
- sequence -
>seq
gcgctagtcgctacgatcgacgctacgtacgatcgatcgatcgatcgatcgatcgactgactagctacgtasgattatacatatattatcgatcga
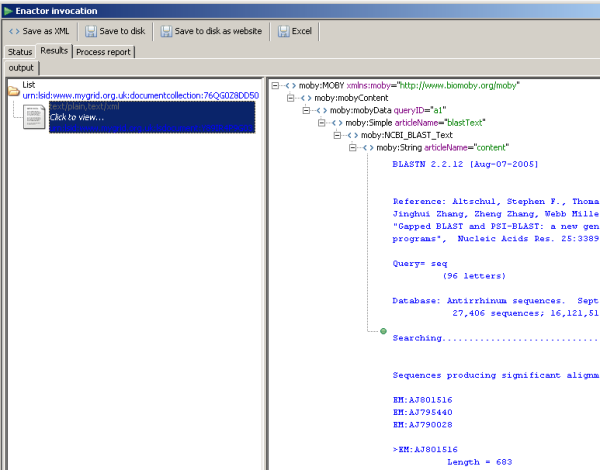
The output from this data is shown below.
The output contains the full Moby message. For details on this messaging structure, click here.
Obtaining information on a Moby datatype
One of the neatest things added to the plugin was the ability to obtain information regarding BioMoby datatypes. For
instance, you can find out what services a particular datatype can be used as input or output. In addition, you can find
the descriptions of those services.As an example, imagine that we wanted to discover what services the Moby datatype 'FASTA' feeds into. To do this, we
can use our existing workflow. From the 'Advanced model explorer' window, context click on the processor called 'FASTA'
and choose 'Moby Object Details'.
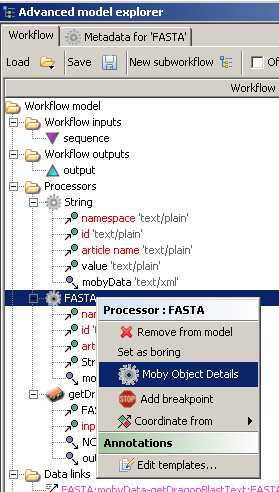
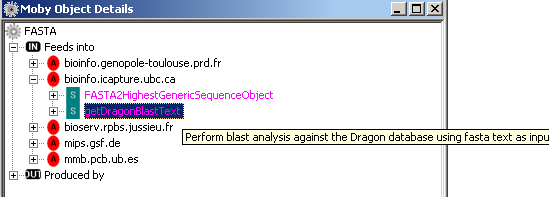
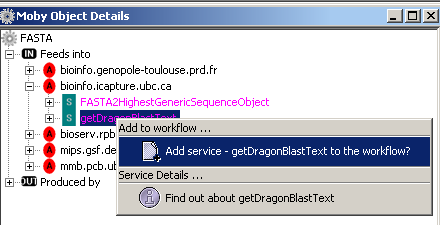
As soon as you choose 'Moby Object Details', a pop up window appears and embedded in that window
is a tree like structure. Note that this window can take up to a minute to appear depending on your
internet connection and the data type chosen. At the root of the tree is the datatype that we chose to
know more about, in our case 'FASTA', and 2 sub trees labeled 'Feeds into' and 'Produced by'. In the
'Feeds into' sub tree, you will find out what BioMoby services consume the datatype in question, sorted by
authority.When you navigate the sub tree and mouse over the service name, a tool tip appears giving you the
description of the service.
In addition, you can context click the service name, in this case getDragonBlastText, and a menu is shown:
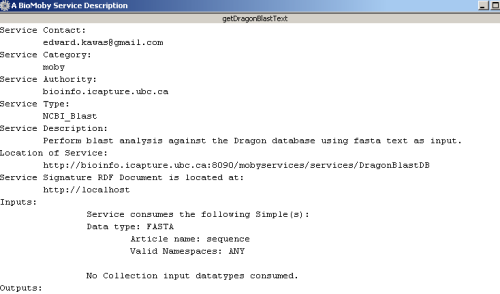
From this menu, you can choose to either add the selected service to the workflow or view more a more detailed
service description. Below, the window that appears upon choosing to 'Find out about getDragonBlastText':
The sub tree 'Produced by' offers similar information, except about services that produce the datatype in question.
The context menu is not available for children of the 'Produced by' node.
Obtaining information on a Moby service
As with Moby datatypes, information about services is also available. For a particular service, we can determine
what are the inputs and outputs. And if you wish, you may add the input datatypes to the workflow.In addition,
we can also discover what namespaces a particular input or output is valid in.Using the workflow that we created above, we can discover what inputs and outputs 'getDragonBlastText' utilize. To determine this,
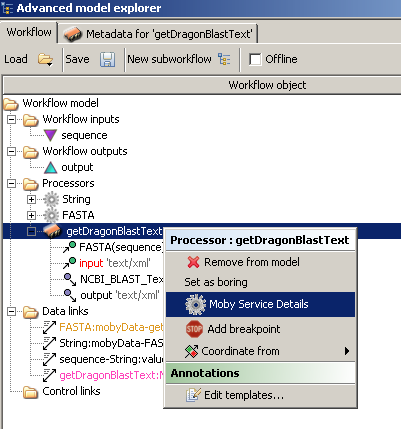
we need to context click on the 'getDragonBlastText' processor from the 'Advanced model explorer', just as we had done before
with the Moby datatype processors above, and choose 'Moby Service Details'.
A pop window appears and embedded in it is a tree that has as its root the name of the Moby
service. The children of the root node are 2 sub trees, 'Inputs' and 'Outputs'. The items under
these sub trees simply illustrate what datatypes the service consumes and produces.
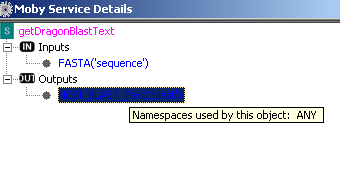
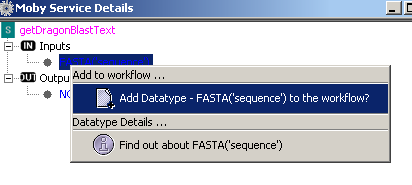
Similarly to the 'Moby Object Details', you can either add the input datatype to the workflow or
obtain additional information on the datatype:
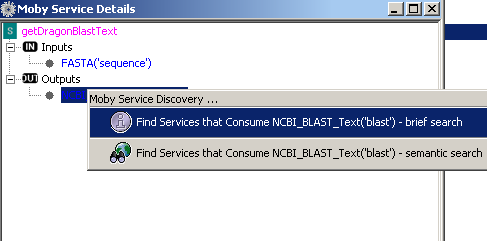
In addition to adding the inputs for a service, you can also determine what other BioMoby services
consume the object that your service, in this case 'getDragonBlastText', produces. This can be done by context-
clicking on the output datatype and choosing one of the following:
- Find Services that Consume NCBI_BLAST_Text('blast') - brief search:
Searches for all services that consume NCBI_BLAST_TextFind Services that Consume NCBI_BLAST_Text('blast') - semantic search Searches for all services that either consume NCBI_BLAST_Text or it parent datatype. For more infomation on this behaviour, click here.
Upon clicking one of the 2 choices, a window similar to the one described above called Moby Object Details
is shown. From this window, you can add to your workflow services that consume, directly or indirectly,
NCBI_BLAST_Text.
Using secondary parameters
Secondary parameters are those that are required in addition to a Primary data inputs in order to modify
the behaviour of the service.If a particular BioMoby service is configurable, you can configure it quite easily. To illustrate how easy it
is to configure Moby services, we will configure a service under the authority, genome.imim.es, named
runRepeatMasker. Adding this service to the workflow, we end up with the following:
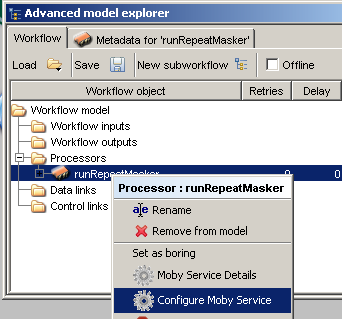
To configure the service, context click on the processor from the 'Advanced model explorer' and choose
the option 'Configure Moby Service':
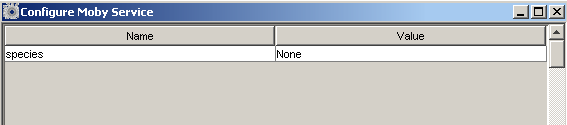
A window will appear with all of the configurable parameters. In this services' case, the only configurable
parameter is the parameter called 'species', with a default value of 'None'.
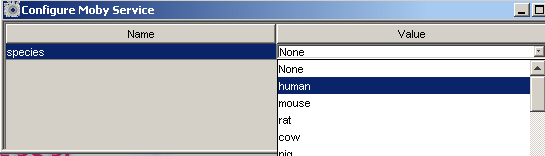
Clicking on the value, will bring up a drop down list of values that we can choose. Once you have chosen
the value you want, close the window and the service is configured.
That's all there is to configuring BioMoby services!
Obtaining the 'data' from a Moby service
Those of you familiar with the Moby plugin since the early beginings of Taverna will recall that you had to
use a local widget for parsing Moby service responses. For simple responses, this was sufficient. For
more complicated responses, this widget became more complicated to use and sometimes resulted in users
having to introduce intermediate services to perform the bulk of the parsing before using the local widget.You should no longer require the use of the local widget 'parse_moby_data' and instead use the method of
parsing service responses outlined below.To illustrate how to parse Moby service responses, we are going to demonstrate how to obtain the data
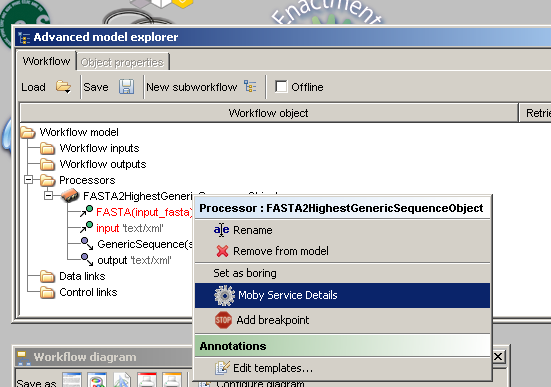
from the service 'FASTA2HighestGenericSequenceObject' under the authority 'bioinfo.icapture.ubc.ca'.Assuming that you have added the above mentioned service to the workflow, context clicking on the service
from the 'Advanced model explorer' will reveal a menu.
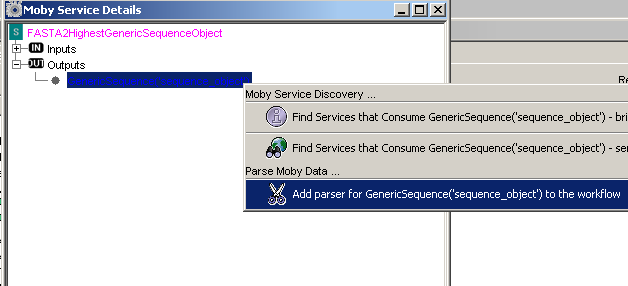
If you select the menu item 'Moby Service Details', a window showing a tree describing the services' inputs
and outputs is shown. Clicking on the node 'Outputs' reveals that this service outputs a Moby datatype
called GenericSequence. Context clicking 'GenericSequence('sequence_object'), reveals another menu that,
among other choices, has the choice to add a parser for this datatype.
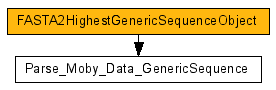
Once you select to add the parser to the workflow, the parser is added to the workflow and correctly linked to
the service output that triggered this action.
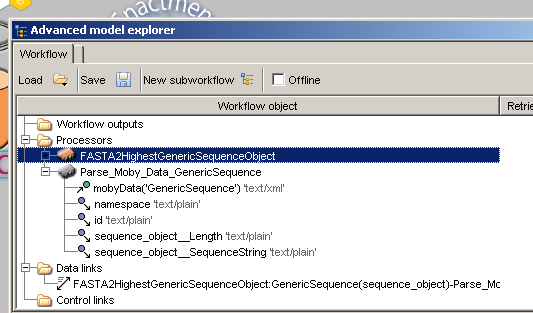
Now, with this parsing processor, you will be able to extract any of the data from the output of
'FASTA2HighestGenericSequenceObject' using the parsers output ports.
What's Next
In the future, we hope to add even more functionality to the plugin. Below are some of the features that
we have thought about:
- Adding the ability to add all services from a particular authority to a workflow.
- Automatic workflow creation given an input datatype and a desired output datatype.
If there are specific features that you would like us to consider, please don't hesitate to contact the BioMoby
development team.
FAQ
1. What are the Moby Service Ports 'input' and 'output' used for?2. What happens if I connect the output port, x, from a Moby service
to the input port, y, of another Moby service?
3. Why do some services have have '_ANON_' or 'Collection - MobyCollection' in there name?
4. If all I want to do is run a Moby service, do I have to specify article names?
5. If I want to connect a service that produces only a 'simple' to a service that consumes a collection,
how can I consolidate all of the simples into a collection?
- What are the Moby Service Ports 'input' and 'output' used for?
Those ports are used to maintain legacy support for users of Taverna prior to version 1.2. When creating new workflows,
you should not be using those ports.
- What happens if I connect the output port, x, from a Moby service to the input port, y, of another Moby service?
There are 3 different types of output ports that Moby services utilize and 2 different types of input ports (excluding the legacy ports
'input' and 'output'.
Moby Service Output ----Connects to ---> Moby Service Input 1 Simple Simple 2 Simple Collection 3 Collection Simple 4 Collection Collection 5 CollectionAsSimples Simples 6 CollectionAsSimples CollectionA Simple is a datatype that is not contained within a collection, i.e. do not have a name that contains '(Collection -'.
A CollectionAsSimples is an output port that contains a Taverna style List of the individuals in a Moby Collection.
Cases 1 and 4 are trivial, because the data in the output port is passed along to the input port without being modified.
Case 2 involves the passing of a Simple, or a list of Simples, to a service that requires a collection of Simples. In this case,
a collection is created containing the simple(s) passed into this service.
Case 3 entails a collection being passed to a Moby service that expects a single Simple datatype. Here, the collection
is broken up into it individual Simple components and passed to the service one at a time. For a collection with n items
in it, the input Moby service is invoked n times, once for each item in the collection.
Case 5 is trivial as well, since this output port contains a Taverna style List of the individual Simple components of a Moby Collection.
This is the prefered way of passing a collection of items to a downstream Moby service that is not expecting a Moby Collection.
Case 6 is very similar to case 2. - Why do some services have have '_ANON_' or 'Collection - MobyCollection' in there name?
These are names automatically generated for services that incorrectly registered with a Mobycentral registry.
You should attempt to contact the service provider so that they may fix and proplerly register these services.
- If all I want to do is run a Moby service, do I have to specify article names?
No.Article names are only required if you plan on using the datatypes for things other than as input to a Moby service.
- If I want to connect a service that produces only a 'simple' to a service that consumes a collection,
how can I consolidate all of the simples into a collection?
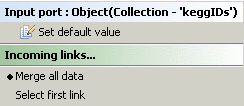
To do this, select the input port of the service that will consume the simples from the 'Advanced Model Explorer' and context click the port.
You will be shown, among other items, the choices to 'Select first link' or to 'Merge all data'.
Choose to 'Merge all data'.
Comments / Questions
If you have any questions or comments, please feel free to contact Eddie Kawas @
If you have any suggestions regarding new features or you would like to report a bug,
please let me know.