try {
MobyPackage mobyInput = MobyPackage.createFromXML (data);
// here do something with 'mobyInput'
} catch (MobyException e) {
System.out.println (e.getMessage());
}
To bring data back into XML is again simple:
try {
// fill response data into a mobyOutput...
MobyPackage mobyOutput = new MobyPackage();
...
// and convert it into XML
String xml = mobyOutput.toXML();
} catch (MobyException e) {
System.out.println (e.getMessage());
}
On the other hand, what is visible to the service developers,
and what they are working with often, are the Java objects created by
the parser. This is a high-level overview of them, and how they
correspond to the Biomoby XML tags:
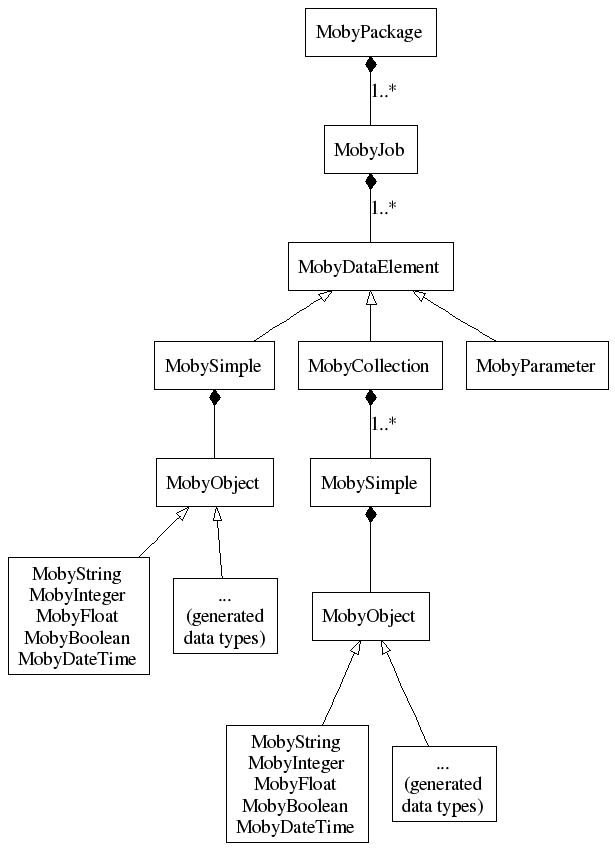
A MobyPackage contains one or more MobyJobs each of them containing one "execution" (or a "query" in a Biomoby speak). Note that a Biomoby service must be able to accept more invocations (jobs, queries) from one network request. A MobyJob also carries a response from a service.
Biomoby XML tag Java object MOBY, mobyContent org.biomoby.shared.parser.MobyPackage mobyData org.biomoby.shared.parser.MobyJob Simple org.biomoby.shared.parser.MobySimple Collection org.biomoby.shared.parser.MobyCollection Parameter org.biomoby.shared.parser.MobyParameter Object and its children org.biomoby.shared.datatypes.MobyObject
org.biomoby.shared.datatypes.MobyString
org.biomoby.shared.datatypes.MobyInteger
org.biomoby.shared.datatypes.MobyFloat
org.biomoby.shared.datatypes.MobyBoolean
org.biomoby.shared.datatypes.MobyDateTime
and all generated data types in org.biomoby.shared.datatypes.*CrossReference org.biomoby.shared.datatypes.MobyXref ProvisionInformation org.biomoby.shared.data.MobyProvisionInfo
A MobyJob has the data themselves - they are either stored in MobySimples or in MobyCollections. The number and types of these MobyDataElements are defined for each Biomoby service in a Biomoby registry.
The MobySimples have data stored in various data objects - their classes are in the package org.biomoby.shared.datatypes. The primitive data types are there available directly, the other types are to be generated - see details in a separate document.
Here is a picture showing how the entities are relates:
The program can be invoked using a script:
At the moment, there is no Ant's target to invoke this program.build/run/run-moby-parser [<options>] <filename>
The program takes the following options, specific for generating data types:
Option/Parameter Ant's property Meaning <filename> XML file to be parsed (a mandatory parameter) -n it does not print the parsed result (by default it does) -r convert back (reverse) the parsed result into XML and print it
The reverse converting (with the -r option) has an interesting usage. The resulting XML is not usually identical with the original - it may have different formatting, it probably has more XML namespace prefixes (they are everywhere), but more importantly, it may not reflect all data that was in the original input. This is because the parser ignores all values that are not carried by (and only by) the Biomoby primitives types (as it was allowed before big change in summer 2005). So you can use this option to see if your XML data are up-to-date with the Biomoby API.
Here is a typical examples of invoking the script - the input file contains an example taken from the Biomoby API documentation:
build/run/run-moby-parser -r example.xml
Authority: http://www.tempcalculator.org/meltyman
Jobs (invocations):
(1) Query ID: a1
Data elements:
(Simple) Article name:
MobyFloat, Id: 163483, Namespace: NCBI_gi
Value: 69.8
(2) Query ID: a2
Data elements:
(Simple) Article name:
MobyFloat, Id: 635543, Namespace: NCBI_gi
Value: 72.1
<?xml version="1.0" encoding="UTF-8"?>
<moby:MOBY xmlns:moby="http://www.biomoby.org/moby">
<moby:mobyContent moby:authority="http://www.tempcalculator.org/meltyman">
<moby:mobyData moby:queryID="a1">
<moby:Simple>
<moby:Float moby:id="163483" moby:namespace="NCBI_gi">69.8</moby:Float>
</moby:Simple>
</moby:mobyData>
<moby:mobyData moby:queryID="a2">
<moby:Simple>
<moby:Float moby:id="635543" moby:namespace="NCBI_gi">72.1</moby:Float>
</moby:Simple>
</moby:mobyData>
</moby:mobyContent>
</moby:MOBY>