Because Biomoby users often ask for examples of Biomoby input data, we created also a simple client that uses the DataTypeTreeTable component outside of the Dashboard. This program is called CreateMobyInput and there are command-line scripts to run it. Be aware, however, of its limitations (they are addresses fully in the Dashboard): no support for collections, and no support for secondary inputs (parameters).
This document describes this client.
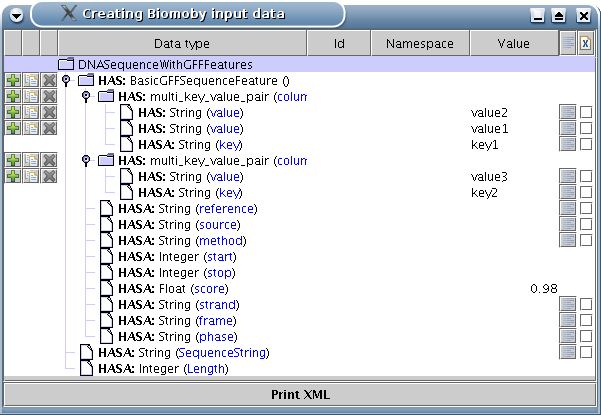
Here is a screen-shot showing the DNASequenceWithGFFFeatures data type with few values already entered, and with few HAS members added:

Clicking on icons in the lines with HAS members allows:
 to add a new (but empty) HAS member
to add a new (but empty) HAS member
 to clone (with all its current values and children) a HAS member
to clone (with all its current values and children) a HAS member
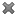 to remove a HAS member (note that if you remove the last one, you
cannot add any back)
to remove a HAS member (note that if you remove the last one, you
cannot add any back)
Additionally, the primitive types have specialized editors for entering values:
 icon. Here is a screen-shot:
icon. Here is a screen-shot:
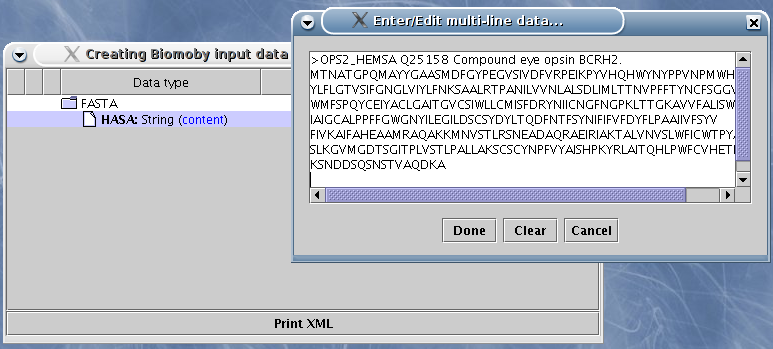
For strings, you can also check the box in the  column - and the string value
will be added to the resulting XML as a CDATA entity.
column - and the string value
will be added to the resulting XML as a CDATA entity.
<?xml version="1.0" encoding="UTF-8"?>
<moby:DNASequenceWithGFFFeatures xmlns:moby="http://www.biomoby.org/moby" moby:id="" moby:namespace="">
<moby:BasicGFFSequenceFeature moby:id="" moby:namespace="">
<moby:multi_key_value_pair moby:id="" moby:namespace="" moby:articleName="column9_tag_value">
<moby:String moby:id="" moby:namespace="" moby:articleName="value">value2</moby:String>
<moby:String moby:id="" moby:namespace="" moby:articleName="value">value1</moby:String>
<moby:String moby:id="" moby:namespace="" moby:articleName="key">key1</moby:String>
</moby:multi_key_value_pair>
<moby:multi_key_value_pair moby:id="" moby:namespace="" moby:articleName="column9_tag_value">
<moby:String moby:id="" moby:namespace="" moby:articleName="value">value3</moby:String>
<moby:String moby:id="" moby:namespace="" moby:articleName="key">key2</moby:String>
</moby:multi_key_value_pair>
<moby:Float moby:id="" moby:namespace="" moby:articleName="score">0.98</moby:Float>
</moby:BasicGFFSequenceFeature>
</moby:DNASequenceWithGFFFeatures>
Once you have jMoby on your computer, you can start it by typing:
build/run/run-create-input -help
Usage:
run-create-input -h[elp]
run-create-input -data <data-type> [<moby-arguments>] [<other-arguments>]
run-create-input -service <service> [<moby-arguments>] [<other-arguments>]
where
-data <gdata-type-name>
specifyies for which data type an input should be created
(e.g. -data FASTA)
-service <gservice-name>
specifyies for which service an input should be created
(e.g. -service Mabuhay)
One of these parameters must be specified.
where <moby-arguments> specify what Moby registry to connect to
-cachedir <dir>
<dir> is a directory with cached entities of a Moby registry
-e <URL>
<URL> is an endpoint of a Moby registry
(default: http://mobycentral.icapture.ubc.ca/cgi-bin/MOBY05/mobycentral.pl)
-uri <namespace>
<namespace> is a URI defining Moby registry's namespace
(default: http://mobycentral.icapture.ubc.ca/MOBY/Central)
where <other-arguments> are:
-q ... quiet mode (prints only errors)
(default) ... prints events what is happenning (to a log)
-v ... verbose mode (prints also decorations)
There are really only two important options: One (even mandatory) is
ether -data or -service. The former names a data
type for which you wish to create input data, the latter gives a
service name for the same purpose. By the way, in order to find what
data types and/or service names are available you can use another
jMoby (command-line) client:
See more details here.build/run/run-cmdline-client -ld build/run/run-cmdline-client -ls
The other important option -cachedir is not mandatory but without using local cache (of Biomoby data types definitions from a Biomoby registry) it will take ages to start. So it is very recommended.
You can find more about how to create such cache in advance using a CacheRegistryClient. But you do not need to - once a -cachedir option is used the cache is created (so the first time it takes longer) and next time it is simply re-used.