This document explains how, as a service provider, you can publish an EMBOSS program running on your machine as a MOBY service. You can also use this framework if you've written an ACD file for any given command line program. The wrapping approach described here requires no Java coding, just the editing of a configuration file.
For scientific publications using this methdology, please cite this paper
The following should be (pre-)installed:
You need to tell ACDServlet where EMBOSS is located on your system. This is done by modifying the
WEB-INF/web.xml file in the ACDServlet WAR you just downloaded. First, extract the file from the WAR:
Then customize it for 1) the target program and 2) your computer setup, using your favorite text editor: Here's the section relevant to the target program (jar xvf ACDServlet.war WEB-INF/web.xml
vectorstrip in our example), with data to customize in bold red.
<display-name>BioMOBY Web Service (EMBOSS): vectorstrip</display-name>
<servlet>
<servlet-class>ca.ucalgary.services.ACDService</servlet-class>
<servlet-name>vectorstrip</servlet-name>
</servlet>
<servlet-mapping>
<servlet-name>vectorstrip</servlet-name>
<url-pattern>/</url-pattern>
</servlet-mapping>
<!-- To make the MobyService name more descriptive than the EMBOSS program name, which tends to be terse,
fill in a BumpyCaseName for the published service below, which will be more intelligible to end-users. -->
<context-param>
<param-name>mobyServiceName</param-name>
<param-value>TrimSequenceVector</param-value>
</context-param>
<!-- A comma separated list of service inputs, of the form "name:objectType:namespace" with :namespace optional. -->
<!-- For an EMBOSS (ACD-based) program, the input and output parameter names must be the same as those specified
in the ACD file (this is how ACDServlet knows how to unify the MOBY and ACD parameters). -->
<context-param>
<param-name>mobyInput</param-name>
<param-value>sequence:DNASequence,linkera:DNASequence,linkerb:DNASequence</param-value>
</context-param>
<!-- Since we are only dealing with DNA in vectorstrip, set the protein option explicitly,
parameters specified here won't show up as service secondaries (but all other params in the ACD file will). -->
<context-param>
<param-name>embossParams</param-name>
<param-value>vectorfile:N,besthits:Y,outfile:/dev/null</param-value>
</context-param>
<!-- Same caveat as for mobyInput -->
<context-param>
<param-name>mobyOutput</param-name>
<param-value>outseq:DNASequence</param-value>
</context-param>
<!-- A suitable entry from the service type ontology. -->
<context-param>
<param-name>mobyServiceType</param-name>
<param-value>Clipping</param-value>
</context-param>
|
In the same file, there are a few parameters that will be the same for all of the services you decide to wrap:
<!-- A domain name here (i.e. identifies your institution for end-users of the service). -->
<context-param>
<param-name>mobyProviderURI</param-name>
<param-value>moby.ucalgary.ca</param-value>
</context-param>
<context-param>
<param-name>mobyAuthorContact</param-name>
<param-value>gordonp@ucalgary.ca</param-value>
</context-param>
<!-- File system location of where EMBOSS is installed, with the bin, share, etc. directories. -->
<context-param>
<param-name>embossRoot</param-name>
<param-value>/usr/local</param-value>
</context-param>
|
Now, put the updated configuration file back into the servlet WAR. For simplicity of deployment, copy the original WAR to new a file named after the Moby service that will be published (in our example, TrimSequenceVector):
cp ACDServlet.war TrimSequenceVector.war jar uvf TrimSequenceVector.war WEB-INF/web.xml
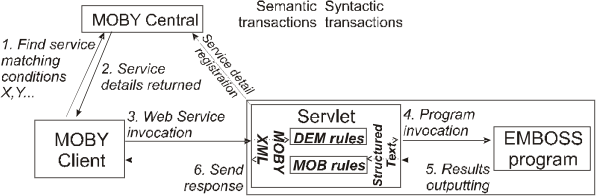
For most EMBOSS programs, no coding will be required, because the ACDServlet WAR has built into it a set of rules that transform the Moby XML messages passed to the service into legacy text formats (e.g. FastA), and vice versa. The general architecture of the service provision is shown below:

If when you test your servlet in Step 4 an exception is thrown, you may be trying to use a Moby
object for which no mapping to the EMBOSS data rtype is available. In this case, you will need to
write either a DEM (XSLT) or MOB (regular expression) rule to do the conversion. Instructions on
how to write these rules can be found here. If you create a new
rule, make it available via a URL and uncomment the corresponding parameter in the web.xml.
We hope to create a Web-based MOB and DEM rule repository in the near future.
Your WAR file is now ready to host your MOBY-S Web Service, and can to be deployed in the Servlet container. How you do this depends on the container. For Tomcat, the easiest way is to use the management Web interface (e.g. http://your.servlet.host:8080/manager/html, but change 8080 appropriately if you had to follow Step 0) and upload the WAR.
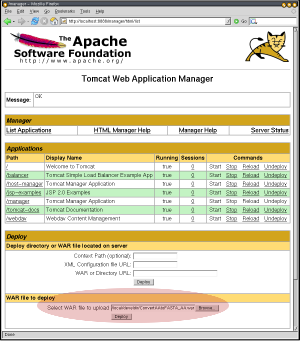 1. Specification of WAR Upload |
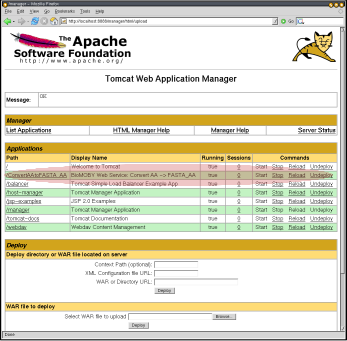 2. Successful deployment |
| Servlet Container | Where to put TrimSequenceVector.war | Further action |
|---|---|---|
| Tomcat | $CATALINA_HOME/webapps | Restart Tomcat |
| WebLogic | $WEBLOGIC_HOME/config/mydomain/applications | Nothing, it loads the WAR automatically |
| JBOSS | $JBOSS_HOME/server/default/deploy | Nothing, it loads the WAR automatically |
You should test your service to make sure it works in the servlet environment. A testing client program is automagically included in your WAR, so type (with the fully qualified host name, and change 8080 appropriately if you had follow Step 0):
If the service fails, a useful error message should be printed to help you diagnose the problem. This would usually be an incorrectjava -jar TrimSequenceVector.war http://your.servlet.host:8080/TrimSequenceVector/ mobyDNASeq.xml
web.xml, the location of EMBOSS was incorrectly specified, or
no data transformation rule exists to create the input or output required (see Step 2).
Finally, if the output looks reasonable (no errors or faults, a real object is returned), you're ready to register your service with MobyCentral so anyone can use it. To do this, simply do the previous command, with an extra argument:
java -jar TrimVectorSequence.war http://your.servlet.host:8080/TrimVectorSequence/ mobyDNASeq.xml registerPermanent
This will run the service to make sure it works, then register it with MOBY Central. You'll see the message "Service Successfully Registered!".
Permanent services: Your service hosts its own RDF specification (retrievable by doing a GET on the servlet URL). An automated agent at MOBY Central retrieves that RDF a few times a day, and if it fails to retrieve it several times in a row, the service is automatically deregistered. Therefore, if you stop or undeploy your servlet for a few days, it will automatically be deregistered. This "passive" deregistration method is for security purposes: no one can (intentionally or accidentally) deregister the service except those who control the servlet container itself.
Syncing with MOBY Central: Your service downloads the MOBY Central ontologies when started, therefore if you want your service to catch an important update to the ontologies, simply restart it (e.g. the "reload" link in the Tomcat manager interface shown previously).
Using a different MOBY Central:
By default, the servlet assumes that the default public Moby Central will be used (this is determined
automagically by a combination of hardcoding and fetching info from biomoby.org). If you want to
use a different Moby Central, e.g. a test registry, uncomment the mobyCentralURL parameter
in the web.xml.